Letter to Editor
The Unique Molecular Structure of Human T-Cell Leukemia Virus Type 1 Protease
Mohsen Karbalaei1 and Masoud Keikha2*
1Department of Microbiology and Virology, Jiroft University of Medical Sciences, Iran
2Department of Microbiology and Virology, Mashhad University of Medical Sciences, Iran
*Corresponding author: Masoud Keikha, Department of Microbiology and
Virology, Mashhad University of Medical Sciences, Mashhad, Iran, Tel: 09386836425; Email:
masoud.keykha90@gmail.com
Submitted: 23 April 2019; Accepted: 29 April 2019; Published: 30 April 2019
Cite this article: Karbalaei M, Keikha M (2019) The Unique Molecular Structure of
Human T-Cell Leukemia Virus Type 1 Protease. JSMC Biochem Mol Res 1: 2.
Human T-cell leukemia virus type 1 (HTLV-1) is retrovirus
type C which causative agent of Human T-cell leukemia virus type
1 associated myelopathy/tropical spastic paraparesis (HAM/
TSP), Adult T-cell leukemia/lymphoma (ATL or ATLL), uveitis,
arthritis, alveolitis/bronchiectasis, dermatitis, and lymphadenitis
[1]. There are approximately 15-20 million people were infected
by HTLV-1 that majority of them where lives in HTLV-1 endemic
area including North America, Central Africa, Caribbean islands,
Japan, Australia and Iran (particularly Khorasan province) [1-2].
Although more than 90% of the HTLV-1 infected individuals are
remains as asymptomatic carrier but 2-6% of HTLV-1infected
individuals are progress to ATLL, as well as 2-3% develop to
HAM/TSP [3].
HTLV-1 is transmitted by unsafe sexually contact, injection,
blood transfusion and breastfeeding throughout population
[2,3]. There is no selective treatment option against HTLV-1;
combination of AZT plus INF-α is only available therapeutic
option of HTLV-1 infection which has not efficient completely [4].
It is suggested that molecular targeting of HTLV-1 has considered
as the best strategy for appropriate treatment; of which, the
Protease enzyme is the suitable option because of its play key
role in process of HTLV-1 polyprotein and has critical role in
HTLV-1 pathogenesis [5]. Therefore, molecular targeting of
HTLV-1 protease has one of the interest option for development
of selective antiviral compounds against HTLV-1 [3,5]. But it’s
necessary to making a survive about structural conformation
and sequence dissimilarities between different strains of
HTLV-1 to design therapeutic option based on the conserved
domains of HTLV-1 Protease which inhibits all strains of HTLV-
1. Therefore, the of this study was comparative analysis and
molecular structure study of the HTLV-1 Protease molecules for
determination of dissimilarities and conserved domains of HTLV-
1 protease for construct of selective HTLV-1 protease inhibitors.
Initially, total of 100 HTLV-1 protease amino acid sequences
of different regions were obtained from NCBI (https://www.
ncbi.nlm.nih.gov/protein); then multiple alignment and the
phylogenic tree was constructed using the neighbor-joining
method with Kimura‐2‐parameter (K2P) distance for evaluation
of genetic diverge among HTLV-1 strains. According to our
results, multiple alignment study (via ClustalW) was showed that
highest similarity in the sequence of HTLV-1 Protease among
different HTLV-1 strains (100-98.93% similarity); also, the
phylogenic tree was approved the majority of similarity in target
gene sequences (Figure 1).
Subsequently, the crystal structure of HTLV-1 protease
was obtained from PDB (3WSJ) for determination of active site
(Figure 2). Then, entirely of the crystal structures of HTLV-1
Protease were obtained from PDB (https://www.rcsb.org/)
and multiple aligned in order to discovery of dissimilarity of the
sequences (Figure 3). We confirmed that amino acid sequence
of the active site of HTLV-1 is nearly conserved complete among
different HTLV-1 strains.
Finally, about 100 crystal structure of HTLV-1 Protease were
superimposed for 3D dimensional and conformation analysis
of the HTLV-1 Protease molecule which showed that a strong
similarity among HTLV-1 Protease structures and confirmed our previous results about homology of HTLV-1 Protease in different
strains (Figure 4).
Overall, HTLV-1 protease is aspartic protease which
responsible for maturation hydrolysis of HTLV-1 polyproteins throughout active site includes two aspartic acid residues
(Asp32, 36), it’s expressed in homodimeric form with each chain
consisting of 125 residues that is highly specific to its substrate
[6]. In recently years, the HTLV-1 Protease is known as the
suitable molecular targets for efficiently treatment of HTLV-1
infection. According to us survive, the active site of the HTLV-1
Protease is conserved among different strains of HTLV-1 and can
be used as target for development of HTLV-1 Protease inhibitors.
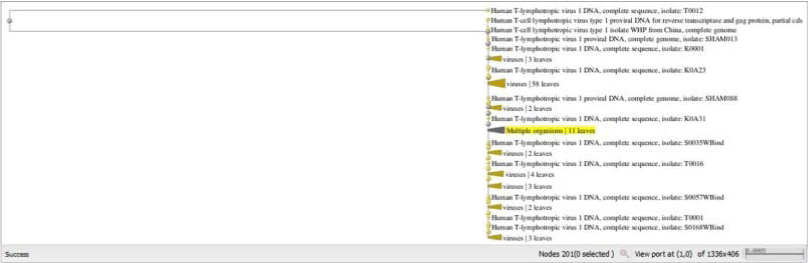 Figure 1:The Phylogenetic tree of the one hundred sequences of HTLV-1 protease using the neighbor joining method. View Figure
Figure 1:The Phylogenetic tree of the one hundred sequences of HTLV-1 protease using the neighbor joining method. View Figure
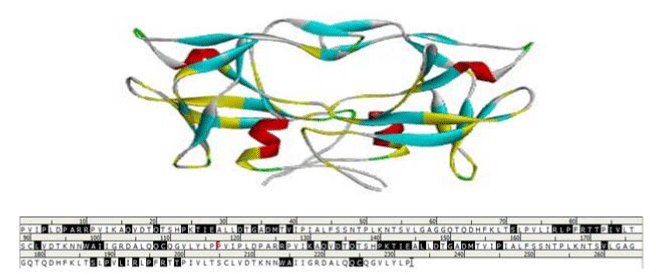 Figure 2:The active site of HTLV-1 Protease was highlighted in the crystal structure and amino acid sequence. View Figure
Figure 2:The active site of HTLV-1 Protease was highlighted in the crystal structure and amino acid sequence. View Figure
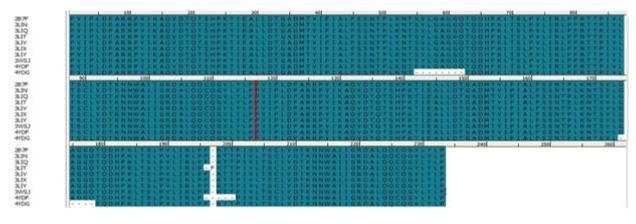 Figure 3:Multiple alignment of HTLV-1 Protease sequences. View Figure
Figure 3:Multiple alignment of HTLV-1 Protease sequences. View Figure
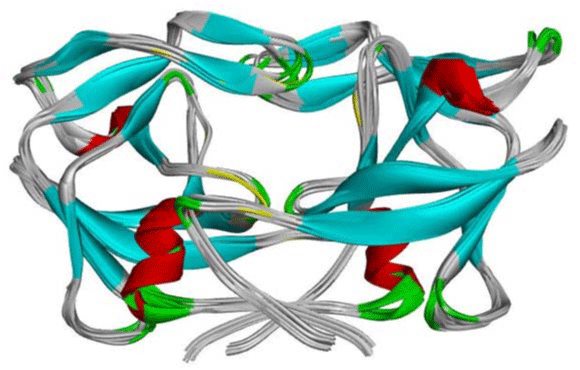 Figure 4:The superimposed of one hundred crystal structure of HTLV-1 Protease. View Figure
Figure 4:The superimposed of one hundred crystal structure of HTLV-1 Protease. View Figure